Difference between revisions of "Explaining My Project"
From GcatWiki
(→Counting Kmers to Tell you about Genome) |
|||
| Line 5: | Line 5: | ||
(taken from https://banana-slug.soe.ucsc.edu/bioinformatic_tools:jellyfish) | (taken from https://banana-slug.soe.ucsc.edu/bioinformatic_tools:jellyfish) | ||
[[File:Kmer.png]] | [[File:Kmer.png]] | ||
| − | Bad kmer rate = bad multiplicity kmers/total number of all kmers | + | |
| − | Seq Error Rate = bad kmer Rate/kmer size | + | *Bad kmer rate = bad multiplicity kmers/total number of all kmers |
| − | Genome Coverage = use gamma fit on the good multiplicity values of the best kmer (usually largest). The peak of this line gives genome coverage (see red line) (here about 47.11x) | + | *Seq Error Rate = bad kmer Rate/kmer size |
| − | Genome size = number of unique good multiplicity kmers/coverage | + | *Genome Coverage = use gamma fit on the good multiplicity values of the best kmer (usually largest). The peak of this line gives genome coverage (see red line) (here about 47.11x) |
| + | *Genome size = number of unique good multiplicity kmers/coverage | ||
Revision as of 19:12, 3 June 2011
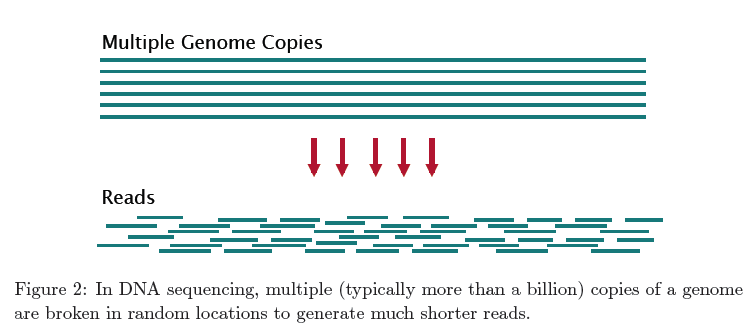
Shotgun Sequencing
Counting Kmers to Tell you about Genome
(taken from https://banana-slug.soe.ucsc.edu/bioinformatic_tools:jellyfish)
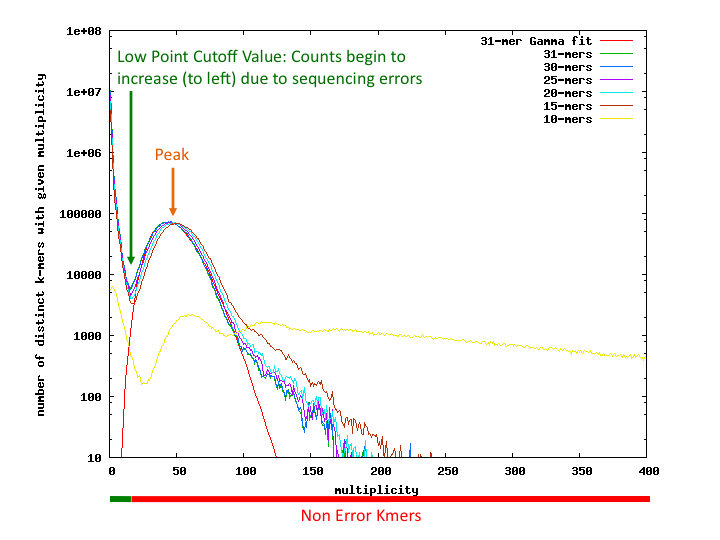
- Bad kmer rate = bad multiplicity kmers/total number of all kmers
- Seq Error Rate = bad kmer Rate/kmer size
- Genome Coverage = use gamma fit on the good multiplicity values of the best kmer (usually largest). The peak of this line gives genome coverage (see red line) (here about 47.11x)
- Genome size = number of unique good multiplicity kmers/coverage