Difference between revisions of "Promoters and Reporters in Synthetic Biology"
(→Synthetic, Artificial, and Mutated Promoters and Reporters) |
(→Synthetic, Artificial, and Mutated Promoters and Reporters) |
||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
[http://en.wikipedia.org/wiki/Promoter Promoters] and [http://en.wikipedia.org/wiki/Reporter_gene reporters] are genetic components used in engineering gene circuits. Promoters are DNA sequences located 'upstream', or ahead, of the DNA sequences encoding genes. Promoters provide binding sites for [http://en.wikipedia.org/wiki/Transcription_factors transcription factors], small proteins that control how and whether DNA is transcribed. Transcription factors bind to promoters in order to give [http://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase] a place to bind to, so that the genes can be transcribed. RNA polymerase binds to DNA and transcribes complimentary RNA from the DNA sequence so that proteins can be formed from the DNA code. If a promoter is being repressed, then transcription cannot occur, as RNA polymerase will not have a place to bind. | [http://en.wikipedia.org/wiki/Promoter Promoters] and [http://en.wikipedia.org/wiki/Reporter_gene reporters] are genetic components used in engineering gene circuits. Promoters are DNA sequences located 'upstream', or ahead, of the DNA sequences encoding genes. Promoters provide binding sites for [http://en.wikipedia.org/wiki/Transcription_factors transcription factors], small proteins that control how and whether DNA is transcribed. Transcription factors bind to promoters in order to give [http://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase] a place to bind to, so that the genes can be transcribed. RNA polymerase binds to DNA and transcribes complimentary RNA from the DNA sequence so that proteins can be formed from the DNA code. If a promoter is being repressed, then transcription cannot occur, as RNA polymerase will not have a place to bind. | ||
| − | Reporters are not as specific as promoters; they are genes that convey some easily-identifiable and measurable characteristic when they are transcribed, such as fluorescence or beta- | + | Reporters are not as specific as promoters; they are genes that convey some easily-identifiable and measurable characteristic when they are transcribed, such as fluorescence or beta-galactosidase proteins. Reporters are generally attached to other gene sequences so the scientist has a way of knowing if the gene is being transcribed - if the reporter is being transcribed, one can assume that the gene of interest is being transcribed as well. |
== Synthetic, Artificial, and Mutated Promoters and Reporters == | == Synthetic, Artificial, and Mutated Promoters and Reporters == | ||
| Line 11: | Line 11: | ||
[http://pubs.acs.org/cgi-bin/article.cgi/achre4/1998/31/i03/html/ar960017f.html Directed evolution] is often used to mutate promoters or reporters in order to obtain desirable attributes. Directed evolution of a gene or protein sequence generally mutates or scrambles the sequence in question, screens it for a certain mutation (any cell not displaying the desirable phenotype is removed), and then amplifies the surviving cells so that the process can begin again. Many mutation and screening cycles can be performed, producing DNA sequences far removed from the original DNA code and increasing the likelyhood that a mutant sequence or cell will have desirable properties. | [http://pubs.acs.org/cgi-bin/article.cgi/achre4/1998/31/i03/html/ar960017f.html Directed evolution] is often used to mutate promoters or reporters in order to obtain desirable attributes. Directed evolution of a gene or protein sequence generally mutates or scrambles the sequence in question, screens it for a certain mutation (any cell not displaying the desirable phenotype is removed), and then amplifies the surviving cells so that the process can begin again. Many mutation and screening cycles can be performed, producing DNA sequences far removed from the original DNA code and increasing the likelyhood that a mutant sequence or cell will have desirable properties. | ||
| − | Another method is the synthesis of combinatorial promoters, as demonstrated in [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html Cox, Surette and Elowitz (2007)]. In their experiment, | + | Another method is the synthesis of combinatorial promoters, as demonstrated in [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html Cox, Surette and Elowitz (2007)]. In their experiment, Cox ''et al''. designed modular sequence units corresponding to the three coding segments of a promoter gene. These segments, assembled at random, can create a diverse and new promoter library made up of fragments of existing promoters, even promoters that are unrelated. See Figure 1 for a diagram of combinatorial promoter synthesis. |
| − | + | In addition, promoters can be specifically synthesized based on the structure of an existing promoter, as in Jensen and Hammer (1997). In order to construct a series of synthetic promoters similar to the ''L. lactis'' promoter, Jensen and Hammer observed consensus sequences within existing ''L. lactis.'' mutants, or sequences that were found to be similar in all or most mutants, no matter how their activity rate varied. For example, the Pribnow box, consisting of the -10 sequence TATAAT and the -35 sequence TTGACA, were consistent in many prokaryotic promoters; other sequences, such as the TG sequence one base pair upstream from the -10 sequence, are more specific to ''L. lactis''. In order to generate a promoter library, Jensen and Hammer constructed oligonucleotides for the sequences that were common in ''L. lactis'' promoters. These oligonucleotides were then seperated by spacers of random sequences; promoters with different spacer sequences made up the promoter library. See Figure 2 for an illustration of the process. | |
| − | |||
| − | In addition, promoters can be specifically synthesized based on the structure of an existing promoter, as in Jensen and Hammer (1997). In order to construct a series of synthetic promoters similar to the ''L. | ||
=== Why use synthetic/mutated promoters and reporters? === | === Why use synthetic/mutated promoters and reporters? === | ||
Since much of synthetic biology is based on modeling genetic and molecular mechanisms before they are built, a scientist has to be able to predict how the components of a mechanism or gene circuit will work in order to predict how the whole mechanism will work. Because they have been specifically designed and selected for, synthetic promoters and reporters make gene circuit modeling much easier. | Since much of synthetic biology is based on modeling genetic and molecular mechanisms before they are built, a scientist has to be able to predict how the components of a mechanism or gene circuit will work in order to predict how the whole mechanism will work. Because they have been specifically designed and selected for, synthetic promoters and reporters make gene circuit modeling much easier. | ||
| − | [http://www.nature.com/msb/journal/v3/n1/full/msb4100185.html Rosenfeld, Young, Alon, Swain, and Elowitz (2007)] have demonstrated that the behavior of a gene circuit can be accurately modeled based on its promoter and repressor activity, but note that in order to accurately construct their model, they needed a specific promoter and repressor gene that followed a | + | [http://www.nature.com/msb/journal/v3/n1/full/msb4100185.html Rosenfeld, Young, Alon, Swain, and Elowitz (2007)] have demonstrated that the behavior of a gene circuit can be accurately modeled based on its promoter and repressor activity, but note that in order to accurately construct their model, they needed a specific promoter and repressor gene that followed a certain pattern of behavior (specifically, a negative regulatory circuit, in which a repressor regulates its own expression, as that circuit is the simplest to model). |
Of course, the noise and randomness inherent in cellular interactions mean that no promoter or reporter's activity can be perfectly predicted. | Of course, the noise and randomness inherent in cellular interactions mean that no promoter or reporter's activity can be perfectly predicted. | ||
| Line 28: | Line 26: | ||
== Measuring, Testing, Tuning, and Modeling Promoters and Reporters == | == Measuring, Testing, Tuning, and Modeling Promoters and Reporters == | ||
| − | * | + | *[[Modeling Promoter Activity | Modeling Promoter Activity: Developing a model for predicting promoter activity based on mutations and gene sequence.]] |
| − | * | + | *[[Modeling Reporter Activity | Modeling Reporter Activity: The kinetics of promoter protein degradation and developing a model thereof.]] |
| − | * | + | *[[Predicting Gene Circuit Activity | Predicting gene circuit activity based on promoter and reporter modeling.]] |
| + | == Figures == | ||
| + | [[Image:Msb4100187-f1.jpg]]<br> | ||
| + | Figure 1. <small>Random assembly ligation generates a diverse promoter library. Promoters can be assembled out of modular sequence units. (A) The assembled sequence of an example promoter. The 5' overhangs of each unit are shown in red. The RNA polymerase boxes (-10 and -35) are highlighted in yellow, and the predicted start site of transcription (+1) is capitalized. Operator colors are consistent throughout the figure. (B) Steps in promoter assembly and ligation into the luciferase reporter vector: promoters are assembled by mixed ligations using 1-bp or 2-bp cohesive ends, and then ligated into a luciferase reporter plasmid. (C) Luminescence measurements in 16 inducer conditions ( each of four inducers, as indicated) for the promoter shown in (A). The output levels determine promoter logic. Note that this promoter does not respond to LuxR regulation at the distal region. (D) The 48 unique units used in the library contain operators responsive to the four TFs (indicated by color) in the regions distal, core, and proximal. </small> In Cox, Surette, and Elowitz 2007. Permission Pending. | ||
| − | + | [[Image:Am0180933001.gif]]<br> | |
| − | + | Figure 2. <small>Strategies used for cloning synthetic promoter fragments into the promoter cloning vector pAK80. (a) Double-stranded DNA fragments carrying putative promoter activities. (b) Restriction map and schematic representation of the relevant parts of the promoter cloning vector. The stippled and solid lines show the strategies used for cloning pCP1 through pCP29 and pCP30 through pCP46, respectively. (c) Restriction map of clones pCP1 through pCP29. (d) Restriction map of clones pCP30 through pCP46. Note that a number of clones have been subject to cloning artifacts and thus may have a slightly different restriction map. BI, BamHI; AII, AflII; Ss, SspI; N, NsiI (PstI compatible); Nr, NruI; Sc, ScaI; HII, HincII; P, PstI; PII, PvuII; E, EcoRI; Sa, SacI; Xh, XhoI; BII, BglII; Sm, SmaI; Xb, XbaI (not drawn to scale).</small> In Jensen and Hammer 1997. Permission Pending. | |
| − | |||
| − | [[Image: | ||
| − | Figure | ||
== Works Cited == | == Works Cited == | ||
| − | |||
| − | |||
*Arnold FH (1997). Design by Directed Evolution. ''Acc. Chem. Res.,''31 (3). Epub 1998 February 28. [http://pubs.acs.org/cgi-bin/article.cgi/achre4/1998/31/i03/html/ar960017f.html Full Text] | *Arnold FH (1997). Design by Directed Evolution. ''Acc. Chem. Res.,''31 (3). Epub 1998 February 28. [http://pubs.acs.org/cgi-bin/article.cgi/achre4/1998/31/i03/html/ar960017f.html Full Text] | ||
*Cox III, RS, Surette MG & Elowitz MB (2007). Programming gene expression with combinatorial promoters. ''Molecular Systems Biology''3(145). Epub 2007 November 13. [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html Full Text] | *Cox III, RS, Surette MG & Elowitz MB (2007). Programming gene expression with combinatorial promoters. ''Molecular Systems Biology''3(145). Epub 2007 November 13. [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html Full Text] | ||
| − | * | + | *De Mey M, Maertens J, Lequeux GJ, Soetaert WK, and Vandamme EJ (2007) Construction and model-based analysis of a promoter library for ''E. coli'': an indispensable tool for metabolic engineering. ''BMC Biotechnology''7(34). Epub 2007 June. |
| + | |||
| + | *Jensen, PR and Hammer, K (1997). The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. ''Applied and Environmental Microbiology''64(1). | ||
| + | |||
| + | *Jensen, PR and Hammer, K (1997). Artificial promoters for metabolic optimization. ''Biotechnology and Bioengineering''58(2-3). | ||
| − | * | + | *Jensen K, Alper H, Fischer C and Stephanopoulos G (2006). Identifying functionally important mutations from phenotypically diverse sequence data. ''Applied and Environmental Microbiology''72(5). |
*Leveau, JHJ and Lindow, SE (2001). Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. ''Journal of Bacteriology''183(23). Epub 2001 September. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=95514 Full text] | *Leveau, JHJ and Lindow, SE (2001). Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. ''Journal of Bacteriology''183(23). Epub 2001 September. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=95514 Full text] | ||
*Miller WG, Brandl MT, Quinones B, and Lindow SE (2001). Biological sensor for sucrose availability: relative sensitivities of various reporter genes. ''Applied Environmental Microbiology''67(3). | *Miller WG, Brandl MT, Quinones B, and Lindow SE (2001). Biological sensor for sucrose availability: relative sensitivities of various reporter genes. ''Applied Environmental Microbiology''67(3). | ||
| + | |||
| + | *Patterson GH, Knobel SM, Sharif WD, Kain SR, and Piston DW (1997). Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. ''Biophysical Journal'' 73. Epub 1998. [http://www.biophysj.org/cgi/content/abstract/73/5/2782 Abstract] | ||
| + | |||
| + | *Rosenfeld N, Young JW, Alon U, Swain PS, and Elowitz MB (2007). Accurate prediction of gene feedback circuit behavior from component properties. ''Molecular Systems Biology''3(143). Epub 2007 November 13. [http://www.nature.com/msb/journal/v3/n1/full/msb4100185.html Full Text] | ||
| + | |||
| + | * Weiss R, Basu S, Hooshangi S, Kalmbach A, Karig D, Mehreja R, and Netravali I (2003). Genetic circuit building blocks for cellular computation, communications, and signal processing. Natural Computing 2 (1). Epub 2004 November 02. [http://www.springerlink.com/content/h885l73711912672/ Abstract] | ||
Latest revision as of 16:37, 10 December 2007
Contents
What Are Promoters and Reporters?
Promoters and reporters are genetic components used in engineering gene circuits. Promoters are DNA sequences located 'upstream', or ahead, of the DNA sequences encoding genes. Promoters provide binding sites for transcription factors, small proteins that control how and whether DNA is transcribed. Transcription factors bind to promoters in order to give RNA polymerase a place to bind to, so that the genes can be transcribed. RNA polymerase binds to DNA and transcribes complimentary RNA from the DNA sequence so that proteins can be formed from the DNA code. If a promoter is being repressed, then transcription cannot occur, as RNA polymerase will not have a place to bind.
Reporters are not as specific as promoters; they are genes that convey some easily-identifiable and measurable characteristic when they are transcribed, such as fluorescence or beta-galactosidase proteins. Reporters are generally attached to other gene sequences so the scientist has a way of knowing if the gene is being transcribed - if the reporter is being transcribed, one can assume that the gene of interest is being transcribed as well.
Synthetic, Artificial, and Mutated Promoters and Reporters
Directed evolution is often used to mutate promoters or reporters in order to obtain desirable attributes. Directed evolution of a gene or protein sequence generally mutates or scrambles the sequence in question, screens it for a certain mutation (any cell not displaying the desirable phenotype is removed), and then amplifies the surviving cells so that the process can begin again. Many mutation and screening cycles can be performed, producing DNA sequences far removed from the original DNA code and increasing the likelyhood that a mutant sequence or cell will have desirable properties.
Another method is the synthesis of combinatorial promoters, as demonstrated in Cox, Surette and Elowitz (2007). In their experiment, Cox et al. designed modular sequence units corresponding to the three coding segments of a promoter gene. These segments, assembled at random, can create a diverse and new promoter library made up of fragments of existing promoters, even promoters that are unrelated. See Figure 1 for a diagram of combinatorial promoter synthesis.
In addition, promoters can be specifically synthesized based on the structure of an existing promoter, as in Jensen and Hammer (1997). In order to construct a series of synthetic promoters similar to the L. lactis promoter, Jensen and Hammer observed consensus sequences within existing L. lactis. mutants, or sequences that were found to be similar in all or most mutants, no matter how their activity rate varied. For example, the Pribnow box, consisting of the -10 sequence TATAAT and the -35 sequence TTGACA, were consistent in many prokaryotic promoters; other sequences, such as the TG sequence one base pair upstream from the -10 sequence, are more specific to L. lactis. In order to generate a promoter library, Jensen and Hammer constructed oligonucleotides for the sequences that were common in L. lactis promoters. These oligonucleotides were then seperated by spacers of random sequences; promoters with different spacer sequences made up the promoter library. See Figure 2 for an illustration of the process.
Why use synthetic/mutated promoters and reporters?
Since much of synthetic biology is based on modeling genetic and molecular mechanisms before they are built, a scientist has to be able to predict how the components of a mechanism or gene circuit will work in order to predict how the whole mechanism will work. Because they have been specifically designed and selected for, synthetic promoters and reporters make gene circuit modeling much easier.
Rosenfeld, Young, Alon, Swain, and Elowitz (2007) have demonstrated that the behavior of a gene circuit can be accurately modeled based on its promoter and repressor activity, but note that in order to accurately construct their model, they needed a specific promoter and repressor gene that followed a certain pattern of behavior (specifically, a negative regulatory circuit, in which a repressor regulates its own expression, as that circuit is the simplest to model).
Of course, the noise and randomness inherent in cellular interactions mean that no promoter or reporter's activity can be perfectly predicted.
Also, synthetic promoters and reporters are useful for when a wild-type promoter or reporter is not sufficient or lacks some property necessary for a cellular mechanism to work. For example, a reporter protein such as GFP does not degrade as soon as it is produced, so in any mechanism that has to detect a transient signal, GFP would not be a useful reporter. However, a mutated GFP, which degrades faster or in the presence of a certain compound, would negate this effect. The same principle applies for reporters which are more active at lower-than-normal or higher-than-normal temperatures. See Patterson GH et al (1997).
Measuring, Testing, Tuning, and Modeling Promoters and Reporters
Figures
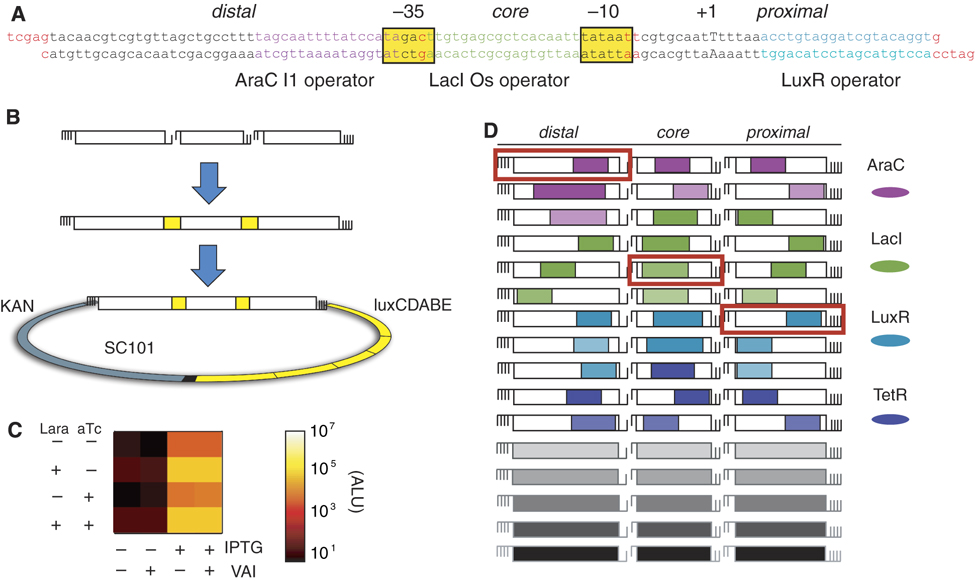
Figure 1. Random assembly ligation generates a diverse promoter library. Promoters can be assembled out of modular sequence units. (A) The assembled sequence of an example promoter. The 5' overhangs of each unit are shown in red. The RNA polymerase boxes (-10 and -35) are highlighted in yellow, and the predicted start site of transcription (+1) is capitalized. Operator colors are consistent throughout the figure. (B) Steps in promoter assembly and ligation into the luciferase reporter vector: promoters are assembled by mixed ligations using 1-bp or 2-bp cohesive ends, and then ligated into a luciferase reporter plasmid. (C) Luminescence measurements in 16 inducer conditions ( each of four inducers, as indicated) for the promoter shown in (A). The output levels determine promoter logic. Note that this promoter does not respond to LuxR regulation at the distal region. (D) The 48 unique units used in the library contain operators responsive to the four TFs (indicated by color) in the regions distal, core, and proximal. In Cox, Surette, and Elowitz 2007. Permission Pending.
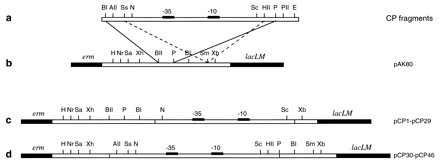
Figure 2. Strategies used for cloning synthetic promoter fragments into the promoter cloning vector pAK80. (a) Double-stranded DNA fragments carrying putative promoter activities. (b) Restriction map and schematic representation of the relevant parts of the promoter cloning vector. The stippled and solid lines show the strategies used for cloning pCP1 through pCP29 and pCP30 through pCP46, respectively. (c) Restriction map of clones pCP1 through pCP29. (d) Restriction map of clones pCP30 through pCP46. Note that a number of clones have been subject to cloning artifacts and thus may have a slightly different restriction map. BI, BamHI; AII, AflII; Ss, SspI; N, NsiI (PstI compatible); Nr, NruI; Sc, ScaI; HII, HincII; P, PstI; PII, PvuII; E, EcoRI; Sa, SacI; Xh, XhoI; BII, BglII; Sm, SmaI; Xb, XbaI (not drawn to scale). In Jensen and Hammer 1997. Permission Pending.
Works Cited
- Arnold FH (1997). Design by Directed Evolution. Acc. Chem. Res.,31 (3). Epub 1998 February 28. Full Text
- Cox III, RS, Surette MG & Elowitz MB (2007). Programming gene expression with combinatorial promoters. Molecular Systems Biology3(145). Epub 2007 November 13. Full Text
- De Mey M, Maertens J, Lequeux GJ, Soetaert WK, and Vandamme EJ (2007) Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnology7(34). Epub 2007 June.
- Jensen, PR and Hammer, K (1997). The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Applied and Environmental Microbiology64(1).
- Jensen, PR and Hammer, K (1997). Artificial promoters for metabolic optimization. Biotechnology and Bioengineering58(2-3).
- Jensen K, Alper H, Fischer C and Stephanopoulos G (2006). Identifying functionally important mutations from phenotypically diverse sequence data. Applied and Environmental Microbiology72(5).
- Leveau, JHJ and Lindow, SE (2001). Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. Journal of Bacteriology183(23). Epub 2001 September. Full text
- Miller WG, Brandl MT, Quinones B, and Lindow SE (2001). Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Applied Environmental Microbiology67(3).
- Patterson GH, Knobel SM, Sharif WD, Kain SR, and Piston DW (1997). Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophysical Journal 73. Epub 1998. Abstract
- Rosenfeld N, Young JW, Alon U, Swain PS, and Elowitz MB (2007). Accurate prediction of gene feedback circuit behavior from component properties. Molecular Systems Biology3(143). Epub 2007 November 13. Full Text
- Weiss R, Basu S, Hooshangi S, Kalmbach A, Karig D, Mehreja R, and Netravali I (2003). Genetic circuit building blocks for cellular computation, communications, and signal processing. Natural Computing 2 (1). Epub 2004 November 02. Abstract