| Main Page | Entry Form | Links |
![]()
Kyte-Doolittle Hydropathy Plots
![]()
Table of Contents
![]()
How do hydrophobicity
and hydrophilicity affect protein structure and function?
Protein Overview
Proteins consist of several amino acids held together with peptide bonds. Each amino acid has a different R group (Figure 1). Structures and abbreviations of all 20 amino acids.
Figure 1: Amino Acid
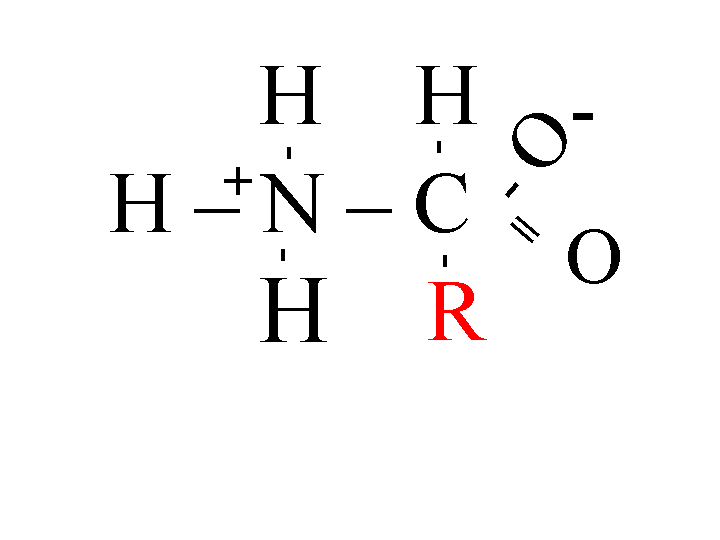
The R group determines whether the protein is hydrophobic or hydrophilic. Hydrophilic groups are typically polar, interacting with water by hydrogen bonding. For this reason, they are called "water loving." Hydrophobic groups, on the other hand, are nonpolar, unable to interact with water, and thus are referred to as "water fearing". The hydrophobicity of the amino acids determines where the amino acid will be located in the final structure of the protein (Kyte, Doolittle 1982). In globular proteins, the hydrophobic R groups will be located on the inside of the protein, away from the water in the cytosol. The hydrophilic R groups will be located on the outside of the protein, interacting with the water in the cytosol (Kyte, Doolittle 1982). An integral membrane protein, on the other hand, must have a stretch of 18-20 hydrophobic amino acids to cross the very hydrophobic inside of the bilipid membrane (Kyte, Doolittle 1982). The hydrophobicity of the inside of the membrane is due to the long hydrocarbon chains of the lipid molecule. All hydrophilic amino acids are pushed to the outside of the membrane. One of the basic tenants of biology is that the structure of a protein defines its function. Being able to make predictions about the structure of proteins will enable biologists to infer more about the protein's function.
Here is an example of a membrane protein.
Here is an example of a globular protein (hemoglobin).
Kyte-Doolittle Hydropathy Plots
Why are Kyte-Doolittle hydropathy plots useful?
Kyte-Doolittle hydropathy plots give you information about the possible structure of a protein. A hydropathy plot can indicate potential transmembrane or surface regions in proteins (Kyte, Doolittle 1982).
How does a Kyte-Doolittle hydropathy plot work?
Kyte-Doolittle plots were first described in a paper by Kyte and Doolittle (1982).
First, each amino acid is given a hydrophobicity score between 4.6 and -4.6. A score of 4.6 is the most hydrophobic and a score of -4.6 is the most hydrophilic. Click here to see each amino acid's score. Then a window size is set. A window size is the number of amino acids whose hydrophobicity scores will be averaged and assigned to the first amino acid in the window. The default window size is 9 amino acids. The computer program starts with the first window of amino acids and calculates the average of all the hydrophobicity scores in that window. Then the computer program moves down one amino acid and calculates the average of all the hydrophobicity scores in the second window. This pattern continues to the end of the protein, computing the average score for each window and assigning it to the first amino acid in the window. The averages are then plotted on a graph. The y axis represents the hydrophobicity scores and the x axis represents the window number.
Setting Parameters and Interpreting Results
By varying the parameters of Kyte-Doolittle tests done on proteins whose structure was known, Kyte and Doolittle(1982) found the parameters that predicted protein structure the best.
When looking for surface regions in a globular protein, a window size of 9 was found to give the best results. Surface regions can be identified as peaks below the mid line. When looking for a transmembrane region in a protein, a window size of 19 is needed. Transmembrane regions are identified by peaks with scores greater than 1.6 using a window size of 19.
What is the GRAVY score?
The GRAVY score is the average hydropathy score for all the amino acids in the protein. It is plotted as a red line on the hydropathy plot. (Kyte, Doolittle 1982). According to Kyte and Doolittle (1982), integral membrane proteins typically have higher GRAVY scores than do globular proteins. Though this score is another helpful piece of information, it cannot reliably predict the structure without the help of hydropathy plots.
Copy and paste the sequences below into the Kyte-Doolittle form. Adjust the window size as you see fit and identify which protein is a globular protein and which one is a transmembrane protein.
Challenge: Identify the transmembrane region(s) of the transmembrane protein.
Hints: Try plotting each sequence with a window size between 5-7 and a window size between 18-21.
Sequence 1
Accession Number: AAA35680 (NCBI Entrez Protein)
1 mqrsplekas vvsklffswt rpilrkgyrq rlelsdiyqi psvdsadnls eklerewdre
61 laskknpkli nalrrcffwr fmfygiflyl gevtkavqpl llgriiasyd pdnkeersia
121 iylgiglcll fivrtlllhp aifglhhigm qmriamfsli ykktlklssr vldkisigql
181 vsllsnnlnk fdeglalahf vwiaplqval lmgliwellq asafcglgfl ivlalfqagl
241 grmmmkyrdq ragkiserlv itsemieniq svkaycweea mekmienlrq telkltrkaa
301 yvryfnssaf ffsgffvvfl svlpyalikg iilrkiftti sfcivlrmav trqfpwavqt
361 wydslgaink iqdflqkqey ktleynlttt evvmenvtaf weegfgelfe kakqnnnnrk
421 tsngddslff snfsllgtpv lkdinfkier gqllavagst gagktsllmm imgelepseg
481 kikhsgrisf csqfswimpg tikeniifgv sydeyryrsv ikacqleedi skfaekdniv
541 lgeggitlsg gqrarislar avykdadlyl ldspfgyldv ltekeifesc vcklmanktr
601 ilvtskmehl kkadkililn egssyfygtf selqnlqpdf ssklmgcdsf dqfsaerrns
661 iltetlhrfs legdapvswt etkkqsfkqt gefgekrkns ilnpinsirk fsivqktplq
721 mngieedsde plerrlslvp dseqgeailp risvistgpt lqarrrqsvl nlmthsvnqg
781 qnihrkttas trkvslapqa nlteldiysr rlsqetglei seeineedlk eclfddmesi
841 pavttwntyl ryitvhksli fvliwclvif laevaaslvv lwllgntplq dkgnsthsrn
901 nsyaviitst ssyyvfyiyv gvadtllamg ffrglplvht litvskilhh kmlhsvlqap
961 mstlntlkag gilnrfskdi ailddllplt ifdfiqllli vigaiavvav lqpyifvatv
1021 pvivafimlr ayflqtsqql kqlesegrsp ifthlvtslk glwtlrafgr qpyfetlfhk
1081 alnlhtanwf lylstlrwfq mriemifvif fiavtfisil ttgegegrvg iiltlamnim
1141 stlqwavnss idvdslmrsv srvfkfidmp tegkptkstk pykngqlskv miienshvkk
1201 ddiwpsggqm tvkdltakyt eggnaileni sfsispgqrv gllgrtgsgk stllsaflrl
1261 lntegeiqid gvswdsitlq qwrkafgvip qkvfifsgtf rknldpyeqw sdqeiwkvad
1321 evglrsvieq fpgkldfvlv dggcvlshgh kqlmclarsv lskakillld epsahldpvt
1381 yqiirrtlkq afadctvilc ehrieamlec qqflvieenk vrqydsiqkl lnerslfrqa
1441 ispsdrvklf phrnsskcks kpqiaalkee teeevqdtrl
Sequence 2
Accession Number: AAA88054 (NCBI Entrez Protein)
1 mvhltpeeks avtalwgkvn vdevggealg rllvvypwtq rffesfgdls tpdavmgnpk
61 vkahgkkvlg afsdglahld nlkgtfatls elhcdklhvd penfrllgnv lvcvpahhfg
121 keftppvqaa yqkvvagvan alahkyh
![]()
Final Note
Kyte-Doolittle plots predict potential protein structures. The graphs produced do not give the final answer about the protein's structure. To increase the reliability of your predictions, remember that what you see depends on the subset range and window size you specify.
Take home message: Be sure to play around with the window size and subset range to increase your predictive power.
![]()
Kyte, J. and Doolittle, R. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105-132.
Last Modified: Wednesday, 27 February 2002 09:04:55 PM