Difference between revisions of "Archaeal BJ1 Virus Hit from Spacer One in Crispr One: Blast Alignments to Other Halophiles"
Karicheson (talk | contribs) |
Karicheson (talk | contribs) |
||
| Line 13: | Line 13: | ||
[[Image:HU1.jpg]] | [[Image:HU1.jpg]] | ||
| + | |||
[[Image:HU2.jpg]] | [[Image:HU2.jpg]] | ||
| + | |||
[[Image:HU3.jpg]] | [[Image:HU3.jpg]] | ||
| + | |||
[[Image:HU4.jpg]] | [[Image:HU4.jpg]] | ||
| Line 24: | Line 27: | ||
[[Image:HS1.jpg]] | [[Image:HS1.jpg]] | ||
| + | |||
[[Image:HS2.jpg]] | [[Image:HS2.jpg]] | ||
| + | |||
[[Image:HS3.jpg]] | [[Image:HS3.jpg]] | ||
| + | |||
[[Image:HS4.jpg]] | [[Image:HS4.jpg]] | ||
Revision as of 03:09, 14 November 2009
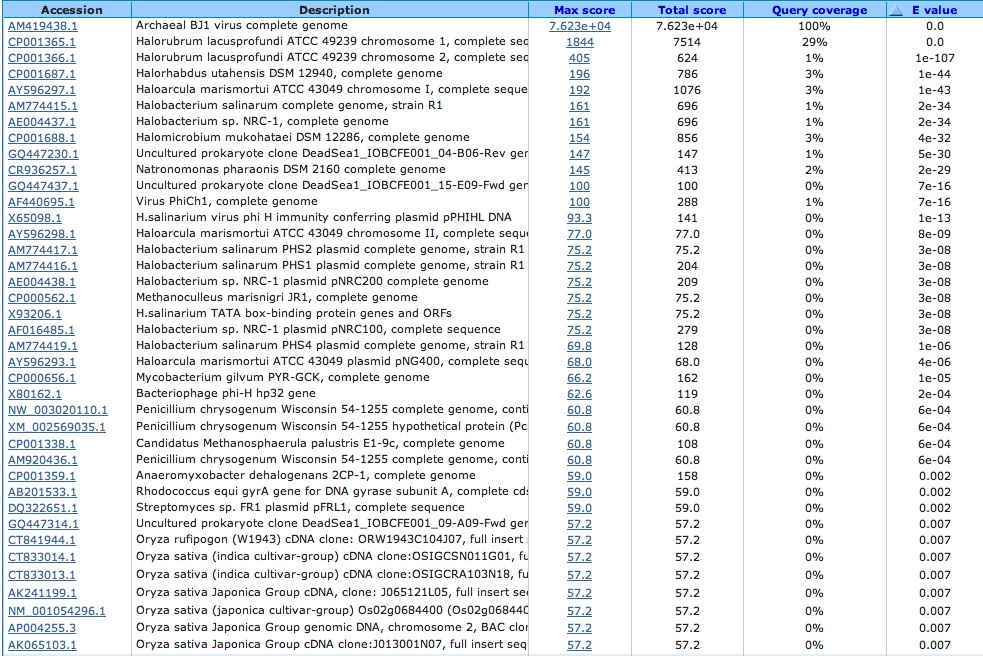
What does this virus primarily attack? (Blasted the virus against nr/nt database)
Infects other halophiles, other bacteria and even other viruses
Blastn Alignment: Query= viral (Accession Number: AM419438.1) and Subject=species of choice
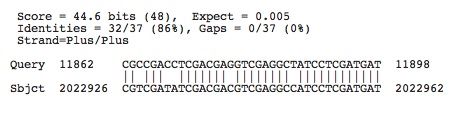
Halorhabdus utahensis: Accession number determined from NCBI: NC_013158.1
Lots of hits, the ones shown below have e-values less than 0.005, the next e-value not included is 0.018
Check to see if any of these hits occur in the spacers of this species by using crispr finder and the nucleotide base numbers
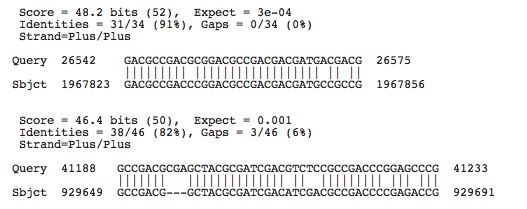
Halobacterium salinarium R1: Accession number determined from NCBI: NC_010364
Lots of hits, the ones shown have e-values less than 0.003, next e-value hit not shown is 0.011
Check to see if any of these hits occur in the spacers of this species by using crispr finder and the nucleotide base numbers
Haloarula californiae
Haloferax volcanii
Haloferax mediterranei
Haloarcula sinaiiensis
Haloarcula vallismortis
Haloarcula marismortui