Mike N
Blueberry Bloom Timing
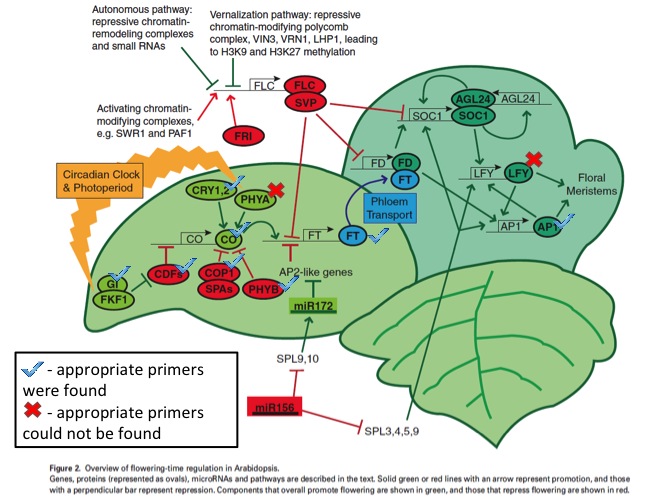
In order to investigate the timing of blooming in blueberries, I found a paper discussing floral timing in Arabidopsis (Amasino, Richard. (2010) Seasonal and Developmental Timing of Flowering. The Plant Journal, 61, 1001-1013.). In this paper, I found a figure describing the known major genes in the blooming pathway (see below, edited).
SSR and Primer
To address some of these major genes I first found their sequences in Arabidopsis. Then, using these sequences as queries, I ran a BLASTn against the blueberry 454 scaffolds on the Genome Database site for blueberry we were given access to ([1]). If the matches were relatively weak, a tBLASTx was also run between the ortholog and database, and the top hits were compared to confirm similarity. I then submitted the scaffold(s) that best match(ed) to the 'SSR finder tool' also on the Genome Database site ([2]). The primers were generated by the 'SSR finder tool' and represent those given that were closest to the location of the orthologs, either bi- or tri- nucleotide repeats with at least 5 repeating units, and that produce a product ranging from 100 to 700 base pairs in length.
EST
In order to see if these genes were actually expressed in blueberry, I submitted sequences near the found genes (often by using areas of the scaffolds where the primers were found) and BLASTed portions of sequences suspected to be near the 3' end of the gene against the blueberry EST database. To confirm whether or not the EST coded tags for my desired genes, I BLASTed the entire sequence of the EST against the entire nucleotide NCBI database ([3]). If the BLAST returned hits that coded homologous or similiar appearing genes, I considered the EST valid.
Paralogs
In order to find potential paralogs within the blueberry genome, I took sequences of ESTs I considered valid and BLASTed them back against the blueberry 454 scaffold database ([4]). As a control I made sure they returned the original scaffold they were produced from, and further investigated the top novel hits to see if they were paralogs. I did this by running an alignment BLAST between the entire scaffold sequence and the orthologous blueberry sequence generated from theArabidopsis sequence. If the sequences aligned well (E-value < .0001) then I considered the hit to be a potential paralog.
Powerpoint
Click Media: Floral_Timing.pptx for a PowerPoint summarizing what I have done, and briefly what I found.
The following is a complete list of what I found for the eleven genes I investigated, including SSR, primer, EST, and paralog data. It also chronicles some of the details of how I obtained my results. Genes are ordered in descending confidence, determined by the E-value of the top hit from the results of the BLASTn between the Arabidopsis seqeunce and orthologous blueberry sequence(and also descending confidence als determined by E-value for the top hits among scaffolds, for genes that had multiple good scaffold hits). The primers are listed in sequential order.
Contents
Crytochrome 1 (CRY1)
SSRs and Primers: yes
ESTs: yes(1)
Paralogs: no
-Scaffold 00331 (@122,000-129,000)
(tc)x5 at 120,300 product size: 251bp forward primer: CATTTTGGGACAGAGGGAGTAG reverse primer: CAGTAACCAACATGCAAAAGGA
When BLASTed against the blueberry EST database, Scaffold00331 returned on EST that is most likely CRY1: EST CV091358
EST CV091358 did not return any apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
-Scaffold 01561 (@18,500-23,800)
(ta)x5 at 11,000 product size: 291bp forward primer: TACCTTAAGGCTCCGTTTGTTT reverse primer: TCCATTTGTTTCGATGTACTGG
(ga)x8 at 37,500 product size: 153bp forward primer: ATCTCCCTACGGTGGGATAAGT reverse primer: AACCTATCGATCCACTCCTTCA
When BLASTed against the blueberry EST database, Scaffold01561 returned on EST that is most likely CRY1: EST CV091358
EST CV091358 did not return any apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
-Scaffold 00649 (@28,300-28,600)
(ga)x6 at 27,600 product size: 245bp forward primer: TTAATTTTGTCCCACCCAAGAC reverse primer: TGAGGGTTCAAAGGACAAAACT
(tc)x5 at 30,800 product size: 162bp forward primer: CTCATTGTCAAACGCAGACTTC reverse primer: TGGTAGTCATCAGGATGGTTTG
When BLASTed against the blueberry EST database, Scaffold00649 returned on EST that is most likely CRY1: EST CV091358
EST CV091358 did not return any apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
Phytochrome A (PHYA)
SSRs and Primers: no
ESTs: no
Paralogs: no
-Scaffold 03861 3,400-5,800
Unfortunately, the scaffold length is only 7,403bp long, so no adequate SSRs, primers, ESTs, or paralogs could be found..
Gigantea (GI)
SSRs and Primers: yes
ESTs: yes (1)
Paralogs: no
Orthologous Arabidopsis sequence from http://www.ncbi.nlm.nih.gov/nuccore/NM_102124.2
BLASTn against 454 database (only good match)
scaffold00100
length=346620
E-value = 3e-51
Section of Scaffold00100 submitted to EST database:
gcttcacgcttgctttttgttgtgttaactgtttgtgtcagtcatgaagc
Only good EST hit:
CV190785
Length = 673
E-value= 3e-33
-Scaffold 00100 (@192,000-200,600)
(ct)x5 at 190,600 product size: 201bp forward primer: ATGATAAAACAATCACCAGCCC reverse primer: GCTTTCGCCTCTACGATCTTTA
(ta)x5 at 207,900 product size: 225bp forward primer: TTTCGCTCAGTTATCTCTCTCTGA reverse primer: CCATCTTTAACTGCACAAACCA
When BLASTed against the blueberry EST database, Scaffold00100 returned on EST that is most likely GI: EST CV190785
EST CV190785 did not return any apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
Cryptochrome 2 (CRY2)
SSRs and Primers: yes
ESTs: yes (1)
Paralogs: no
-Scaffold 00649 (@16,000-30,000) (Note this scaffold was also a hit for CRY1)
(tc)x5 at 30,800 product size: 162bp forward primer: CTCATTGTCAAACGCAGACTTC reverse primer: TGGTAGTCATCAGGATGGTTTG
When BLASTed against the blueberry EST database, Scaffold00331 returned on EST that is most likely CRY2: EST DW043244
EST DW043244 did not return any new (as in other than these two scaffolds) apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
-Scaffold 01561 (@20,000-21,000) (Note this scaffold was also a hit for CRY1)
(ta)x5 at 11,000 product size: 291bp forward primer: TACCTTAAGGCTCCGTTTGTTT reverse primer: TCCATTTGTTTCGATGTACTGG
(ga)x8 at 37,500 product size: 153bp forward primer: ATCTCCCTACGGTGGGATAAGT reverse primer: AACCTATCGATCCACTCCTTCA
When BLASTed against the blueberry EST database, Scaffold01561 returned on EST that is most likely CRY2: EST DW043244
EST DW043244 did not return any new (as in other than these two scaffolds) apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
Phytochrome B (PHYB)
SSRs and Primers: yes
ESTs: no
Paralogs: no
-Scaffold 00751 (@84,000-89,200)
(aga)x8 at 81,900 product size: 265bp forward primer: CCCGAAAATACCCTTTCTCTCT reverse primer: GGCAATTACCAATTACGTGTCA
(ag)x10 at 92,400 product size: 262bp forward primer: AAGAGGGGTAGACCAAAATTGA reverse primer: AATTTCACTCCAACCAAGAAGG
When BLASTed against the blueberry EST database, Scaffold00751 did not return any valid ESTs for PHYB.
No apperent paralogs were found for PHYB in blueberry.
Constans (CO)
SSRs and Primers: yes
ESTs: no
Paralogs: no
Orthologous Arabidopsis sequence from: http://www.ncbi.nlm.nih.gov/nuccore/NM_001036810.1
BLASTn against 454 database (only good match)
Scaffold01843
length=51900
E-value = 2e-19
Top hit with tblastx against 454 scaffolds was also scaffold01843
-Scaffold 01843 (@40,900-41,000)
(ac)x6 at 31,800 product size: 285bp forward primer: CCAAGATCCTTCCAAACTAACG reverse primer: TTCTTCTTCTTCTTCGTTTGCC
(gaa)x5 at 32,000 product size: 133bp forward primer: AGGGGTTAACAAAACATACCCC reverse primer: TCTCTGGTTCAATTTAGGGCTC
(tc)x11 at 48,300 product size: 275bp forward primer: GAAACAGATGGCATGGTGAGTA reverse primer: CTCCAAAACCCTATGAAAGTGC
When BLASTed against the blueberry EST database, Scaffold01843 did not return any valid ESTs for CO.
No apperent paralogs were found for CO in blueberry.
Constitutive Photomorphogenisis 1 (COP1)
SSRs and Primers: yes
ESTs: no
Paralogs: no
-Scaffold 00752 (@17,000-24,800)
(ga)x6 at 14,800 product size: 277bp forward primer: CTCCAACTCTGAACTGATTCCC reverse primer: GAAACGCGTCCTTGATTATCTC
When BLASTed against the blueberry EST database, Scaffold00752 did not return any valid ESTs for COP1.
-Scaffold 00111 (@154,000-162,000)
(ct)x9 at 144,800 product size: 164bp forward primer: GTGGAAAATGTAAAGACACGCA reverse primer: AAAGCTTTCTAACCTCCGATCC
(at)x5 at 165,400 product size: 300bp forward primer: CTTTCTTCCACTCAAGCCCTAA reverse primer: GAATCTTGTGCACCACACACTT
When BLASTed against the blueberry EST database, Scaffold00111 did not return any valid ESTs for COP1.
No apperent paralogs were found for COP1 in blueberry.
Cycling DOF Factor (CDF)
SSRs and Primers: yes
ESTs: yes (1)
Paralogs: yes
-Scaffold 00079 (@69,000-69,200)
(ta)x6 at 61,500 product size: 217bp forward primer: GCTCTCTTTCTCCATGCCTTTA reverse primer: CCCCCAATAACCCTCATTTATT
(aat)x6 at 69,900 product size: 292bp forward primer: GTTTGACATATCGAGCTTGCAC reverse primer: TTGGTTGTAGAGGAGTGGGATT
When BLASTed against the blueberry EST database, Scaffold00079 returned an EST that is most likely CDF: EST DR068043
EST DR068043 did return new apparent paralogous scaffolds when BLASTed back against the Blueberry 454 scaffold database.
-Scaffold 00651 (@19,900-20,100)
(ac)x6 at 14,200 product size: 192bp forward primer: AACTCTTAAAAAGGGACGGAGG reverse primer: CTTTCCCGGTTTCTTGTTTGTA
(ag)x6 at 22,300 product size: 276bp forward primer: CGTCTTCCCAAAGAGCTTAATG reverse primer: TTCTAAGGCCACAAAACCAACT
When BLASTed against the blueberry EST database, Scaffold00651 did not return any valid ESTs for CDF.
-Scaffold 01102 (@51,500-51,700)
(tg)x7 at 45,400 product size: 253bp forward primer: AGGATGAGTGAAAGAGCGTACC reverse primer: CCAGATTTTTCAGAGAATTGGC
(ga)x5 at 66,700 product size: 129bp forward primer: ACCATATTATTGGACCCAGGTG reverse primer: ATAGCACAAACTCCCAAAATGG
When BLASTed against the blueberry EST database, Scaffold01102 did not return any valid ESTs for CDF.
-Scaffold 00292 (@195,200-195,300)
(ac)x8 at 179,200 product size: 282bp forward primer: GATCCATGTTGTTGTGGATTTG reverse primer: TTCTCGAAGATCATTGGAGGTT
(ct)x6 at 197,600 product size: 299bp forward primer: AGCCATTTGAGTTTTCAGGTGT reverse primer: TCACCCTCCACTAGGACTTGTT
When BLASTed against the blueberry EST database, Scaffold006292 did not return any valid ESTs for CDF.
Apetala 1 (AP1)
SSRs and Primers: yes
ESTs: yes (3)
Paralogs: no
-Scaffold 00988 (@71,100-71,200)
(tc)x5 at 70,500 product size: 221bp forward primer: AAGAGCAAGAGACATGACGGAT reverse primer: GTTTTTGGGTCTTGATGGGATA
(tc)x7 at 71,200 product size: 198bp forward primer: CAACCTCAGCATCACAGAGAAG reverse primer: GGGGAAGAATTGAACAACAGAG
When BLASTed against the blueberry EST database, Scaffold00988 returned three ESTs that are most likely AP1: EST CF811608, EST CF810466, and EST CV090688
EST CF811608 returned nine new apparent good hits, but further investigation is needed to determine if they are paralogs.
EST CF810466 returned an additional apparent good hit, but further investigation is needed to determine if it is a paralog.
Flowering Locus T (FT)
SSRs and Primers: yes
ESTs: yes (2)
Paralogs: yes
Orthologous Arabidopsis sequence from http://www.ncbi.nlm.nih.gov/nuccore/GU120195.2
BLASTn agaist 454 database (only good match)
>scaffold00357
length=260075
E-value = 5e-09
Top hit with tBLASTx against 454 scaffolds was also scaffold00357
Section of Scaffold00357 submitted to EST database:
TCACTTTTTAGTGTATAAATACGACTCGATCGCTAGTcTCTCTTGGCTATCTGCAGTAGTAAAAAGCAATAGATATAATCATAAATATATTCTATGCCACGGGATAGGGATCCTCTT GTTGTCGGGCGTGTGATTGGGGATATTTTGGACCCTTTCACAAGATCAATTGGCCTTAGGGTGATATACAATACTAGGGAAGTTAACAATGGGTGTGAACTTAGGCCTTCTCAAGTA GCCATCCAACCAAGGGCTGATATTGGAGGAGAGGACTTAAGGACCTTCTACACTCTTGTAAGTCCCCAATTCACTGTTTTTCTCATCTTTTCCATTAATTCGCGTTCATTATTATAA TTGTTAGTTAAAAAAAAaTGCAAGTAAAAaCAGCtcATTGAACTAATATATGGTTTAAAAaGTTTATTGAAAAGTACAAAAAGTTtAATGgATTATTTGTTCTTTtGTCGGTTGGTG AGTTTTTtttaTAGTTCAGTGAGCTGTCAATATAAAAaTCTTTTTTtCCCCTtGATTTAGAAAAaTGCAAaCAAATATAAGCATTGATATGTCcACcAAAaCGGTTTTTCAATTTGA ATaTCCATTAATTACAACcAAGTAGaTATGTtATTTCATGCAAATCATTCACGGGTATCAGGAGCATTTTCACTTCTTTTCGACACAAACTTAACCTAAGGTTTCTAGTACTTACTA CTTAGATTTATCTGTCATAACAGGTTATGGTGGACCCTGATGCTCCAAGTCcAAGTGACCCCAACCTAaGGGAATACTTGCATTGGTAATGGCCTCTCTCTCCATCTTTCTCTCCAT GGTGTTACCGGGAGctCGGTTgcTTCATAATTAtcTGAATTTAAGTACATTTTTAGAACTTgTTTtgAAaGGAAATGACAATCATCACAAATATaTGTAACTATATATATTATCCAT AATTTCTGTTTTACCATCcAACATCATTTCGAGTTCGAGTTGTTTTtCtCCtAGTCTTTTTtCCTTGATCGATAGATAGAATTCGATTGGTtCCTTCATAATAACCATAACAAaTAT TACAAGAGAGCGGgCAAaTAGAAaGTACGTACAtACcACAAAAaaTtCAATAAATGgAAAAAAAAAAAaaGgaCCAGTtCTcTTGGTATCAAAaCTGCcACCCCACCTCtACAaTaT ATTCTTACATCAAGGAAAAGgAAaCACAAAaGGTAGTACGATGTTGGATGGGAAAATCATTCAAAGCTGGAAGTTAAAACCGATTGTTCAGTTtAcTTTtCATGGGAAAAaGATATG CCTGCATGATCTTATTCtCTTGTTtCTGATTGACTGATTTATGAGTAGACGTGCCTTTCATTGGAACATGCCCACAAGGCCACAACACAAAAGTTCTTTCGATTGAGAGTTTTATGT TTtGTATATGCAACTGTGGCCCCcAACATgTGTACTATGTTTCTGGAAACGTTCCTACTTTTTGAGCCCTTCTTTtCCTCCATTCCCATACTGTTCTTTATTTTACGTACATAAGCT GTCACTCGTTCATTCGATGCATAAAATTCCCcAAAAAATTATGAAATACTATTTTGGCTGAAGTTgAaTTTAACAAAATAAAACACAAATAATTAGGGAAGGAATTTTAAAAaGTTA AATAAGTTAAAAGTGTTATAGGACAAATgATAATTTTGGCTCGATATATGTAGGAACAAAACGTCGATGATCAGTTTtGaGGGGCATTTAAAAAGCACAGTATATAAgaTTAGCATT CgTCCCCTTGGCaTTACAACTTACAAGAGTAAATAaTATCTTTGTAAGACaCaTTCGATTTGTTtGTTtCAACaagCTAGATTTAAGATATTTTAACAGGCTTAATTtGAATAGTGC cgCTAACTGATTCTCGACAAACTAattaaGaCGAcAAATTtCtaCTAAAaaaaatAAAaTTTACACTCCATACAGTtCAACGCTACAGTTATGcACCCGgAGATCGTAATCTAAGCT GCTATTATATAACACTCTCTGGTTGgACATCTTATCTAGGTTGGTGACTGATATTCCAGCAACCACAGGAGCTAGCTTTGGTGAGTAAACtATTTTTCTTCATTTtCCTCCTCagTT GTACATGGAGGgCTCTCTGGTCCCTTTTAGAGACTGGACTGTCCTTTCGGGTATTTTGTGGTCTTTTTCCGGGTCCATTATGATAATCGAAATTGtGCtCTTTTTAAcAgCTTGTAG AAATCAAAATCCAaGTTAAgAAaCATTTTCTTCAAATATTGTTTGATATTTTTACTCTCAGAAAACATCAATGCTATGTAAAATATTACAACACTAAATCGGAGTTCCTTTCTATTA ATAACATAAATGTATTTCAAGGATATACTAATTAATACCCGATAAATACGTTTTTGGCAAGAATATTTTTAGAGAATATTTTTTCAACATTGGGATATATTTTTaCTCTTCCAGTTC TTCTCcTCGAGCcGAATGATGTGATGtATATAaAAaTTCgACcTGAaTCTGgTAAAaGCTAAGAAAaaCAaCAGTAAaGACTGaTAAAAaCAcACTAAAGTTAGTCCAAaGgTaGCT AACCTGgCcTGgTGgTCATATTTTTtAaTtGGTGATCATCATCTATTAGAACTTTTTTGGtGGGTCATTGTAGGACATGAGGTGGTTTGCTATGAAAGTCCACGGCCATCCATGGGG ATTCACCGTTTCGCATTCGTGTTGTTCCGACAaCTGGGCCGCCAAACaGTCTATGCtCCcGGGTGGCGTCAGAATTTCAACACTAGGGACTTTGCCGAGCTTTATAATCTTGGCTCG CCTGTTGCCGCTGTTTATTTTAATTGCCAGAGGGAGAGCGGCTCAGGAGGACGACGACGCTGATACTTACACACATGCGCGTGCATATATTATCTTTGAAAATAATTTTTGCACTCT ACATTCTTATGAACACACTCCAAAAATATTTTGTAGTAGTCTAAATTgTACGCTACACAGTTGTCAAAGTACACTAGAAGATAAATTTtGGAGTA
Top two EST hits:
>CV091156
Length = 779
E-value= 0.0
>CF811337
Length = 669
E-value = 0.0
-Scaffold 00357 (@56,800-58,200)
(ga)x6 at 53,400 product size: 250bp forward primer: TCATTACTCCTTTGCCCATTCT reverse primer: CTTTCTCGGGTGATTCAATGAT
(ag)x5 at 54,300 product size: 130bp forward primer: GATTTCTGGATGGATTCTCAGC reverse primer: ATCCCAAATACAAAACCCACTC
When BLASTed against the blueberry EST database, Scaffold00357 returned on two ESTs that are most likely FT: EST CV091156 and EST CF811337
EST CV091156 returned a new apparent paralogous scaffold, Scaffold01826 when BLASTed back against the Blueberry 454 scaffold database.
EST CF811337 returned an additional new apparent paralogous scaffold, Scaffold00429 when BLASTed back against the Blueberry 454 scaffold database.
Leafy (LFY)
SSRs and Primers: no
ESTs: no
Paralogs: no
Orthologous Arabidopsis sequence from: http://www.ebi.ac.uk/ena/data/view/M91208
No ortholog could be found in Vaccinium when a BLASTn was run in the 454 scaffold database when the Arabidopsis sequence was used as the query. Therfore no SSR, primers, EST, or paralogs could be identified for LFY.